|
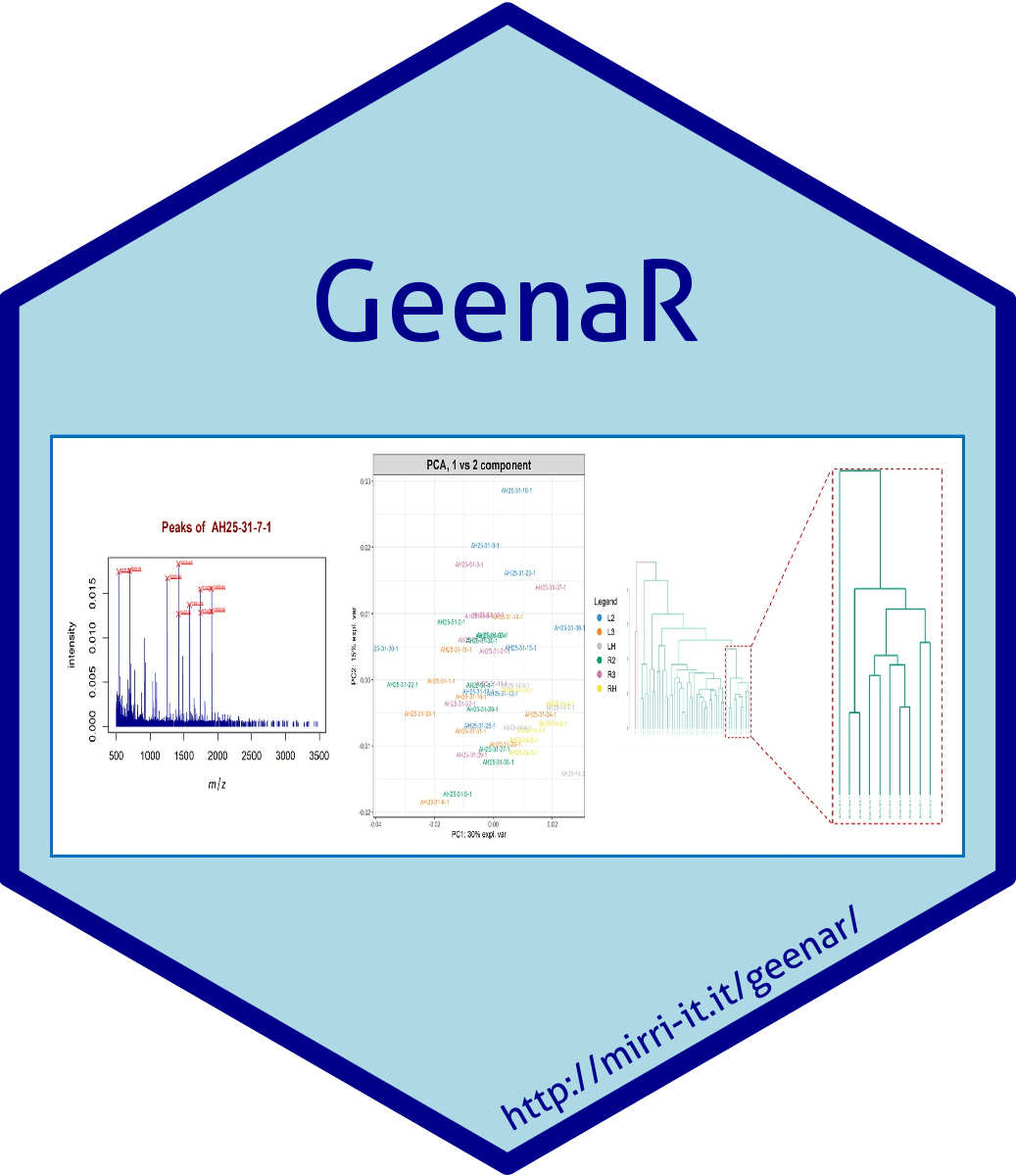
Welcome to GeenaR, a tool for MALDI/ToF MS spectra analysis.
Using GeenaR for analysing MALDI spectra should be straightforward. Note that you cannot use it for LC-MS spectra!
This page may help you in understanding how the system works and using it proficiently.
See also the help page on how to run the test on the example dataset.
|
|
Result page
The output page is composed by three sections, as follows:
- Job summary section: provides a summary of the job, including job and data set names, and steps, methods and parameters of the analysis.
From here, a valid attribute file can be downloaded for later reuse.
- Elaboration section: provides information on the running job. Main steps are listed with starting and ending time.
At the end of the execution, link(s) to report(s) are also provided in this section.
- Results section: provides some of the figures created during the run.
If the user provides a valid email address, a short message with links to the main results is also sent by email.
See an example in the Email message section below.
|
Contextual help
Each field in the input form has a contextual help associated.
You can easily spot it because it is idenfied by the special icon  . .
NB! Move your mouse over the icon  and the contextual help will be shown. You don't need to click on it.
and the contextual help will be shown. You don't need to click on it.
|
This section includes the following fields (mandatory fields are marked by a *):
- Job name: This is a reference name for your analysis.
GeenaR generate a random name that includes a prefix 'Geenar_' and a random three digits number.
The job name, however, can be any sequence of letters, numbers and underscores. So, you may change the name provided by GeenaR at your ease and provide, e.g.i, a label related to your study.
Note that the job name is used internally by GeenaR both as a name for a folder for the run and within the name of result files. Using the same job name of a previous run for a new one will destroy all results previously generated.
NB! If you use the same job name for more than one analysis, results will be overwritten and you will only be able to retrieve them for the last analysis perfromed.
- Data set name: This is the name assigned to your data set when uploading it to the server.
The dataset name may only include letters (both upper- and lowercases), numbers and the following characters: dash, underscore, dot. Avoid spaces!
The data set name is used internally by GeenaR along with the email address for the generation of a folder where the spectra of the data set are stored.
NB! For the example dataset, use 'example' as dataset name.
NB! You can use the same data set name for uploading spectra at different times. All spectra will be included in the same data set and available for later analysis.
NB! At present, you can only analyse spectra which are included in the same dataset. However, spectra can be included in more than one dataset.
- Email: Provide a valid email address. This must correspond to the email address associated with the data set when uploading it. A summary of the run will be sent by email.
NB! For the example dataset, use your email address!
- Country: This is your country. This field is not mandatory. If you provide it, we will update our statistics on GeenaR user countries.
This section includes the following fields (mandatory fields are marked by a *):
This section is divided in four subsections, each of which includes three columns.
Apart from the first subsection, that relates to headers, the three columns includes, respectively from left to right, the name of the step of the analysis, the methods for carrying out that step and the related parameters.
- Headers subsection:
In this subsection, three buttons are included, one per column, controlling the values of fields in the respective column.
In the leftmost column, related to the analysis steps, one checkbox is available for selecting / deselecting all steps.
In the central column, related to methods, a button named 'Set default methods' is available for setting all methods to their default values.
Similarly, in the rightmost column, related to parameters, a button named 'Set default parameters' is available for setting all parameters to their default values.
Note that the reset of all steps, methods and parameters to their default values can also be achieved by clicking on the 'Reset' button of the form.
Also, note that there are dependencies among steps. As a consequence, some steps may or may not be executed depending on the actual execution of some other step.
- Pre-processing subsection:
In this subsection, you may specify which pre-processing steps should be executed on submitted spectra.
The following steps are available:
- Variance stabilization:
This step applies a non-linear transformation on the spectra intensities to reduce the dependency between variance and mean. In particular, the variances of the raw intensities are often a function of the mean intensities. Hence, the variance of the noise is not constant across the spectra making the spectra analysis more challenging. To reduce such an effect, it is possible to apply a non-linear transformation on the intensities to stabilize the variance.
The user can choose one among the following non-linear transformations: square root (SQRT), logarithm base e (LOG), logarithm base 2 (LOG2), logarithm base 10 (LOG10).
These transformations help in the graphical visualization of the spectra and in the handling of the assumptions for using the remaining approaches.
 Tech Hint! Tech Hint!
The transformation step uses the transformIntensity() function
of the MALDiquant package.
- Smoothing:
This step smooths the mass spectra using a convolution with some filters. The approach is typically used in signal processing for denoising.
The user can choose one among the following filters:
- Savitzky-Golay (Savitzky, Golay,1964; Broomba, Ziegler, 1981): The Savitzky-Golay method is a well-known digital filter that smooth a signal by fitting a subset of data points in a given window with a low-degree polynomial by the linear least-squares method. The degree of the polynomial is set to 3. The user can choose the parameter hw that denotes the size of the half window.
- Moving Average: The Moving Average filtering method performs a similar smoothing strategy with the average instead of the local polynomial. The weights in the average are equal. The user can choose the parameter hw that denotes the size of the half window.
The half window size (hw) should be much smaller in the Moving average method than in the Savitzky-Golay method to conserve peak shape. The size of the window is (2*hw + 1).
 Tech Hint! Tech Hint!
The normalization step uses the smoothIntensity() function
of the MALDiquant package.
- Baseline removal:
This step copes with the drift in the signal intensities that typically affect the spectra in a non-linear fashion. The presence of a baseline drift is a commonly encountered problem during the measurement of spectra. It is essential to remove such drift before applying any further analysis without destroying the peaks that characterize the sample. In this task, we first estimate a baseline function (i.e., the baseline drift) using one of the four methods available, then we remove the estimated baseline from the spectra. After such a step, the intensity of the mass spectra would be reduced by the baseline.
The methods available for the estimation are:
- SNIP: it is an iterative method for estimating the baseline based on the approach proposed initially in Ryan et al. (1988),
then improved in Morhac (2009).
- TopHat: it is a method for estimating the baseline based on the algorithm originally proposed in van Herk (1996).
It applies a moving minimum (erosion filter) and, subsequently, a moving maximum (dilation filter) on the intensity values.
- Convex hull: it estimates the baseline using a convex hull constructed below the spectrum, as proposed in Andrew (1979).
- Median: it estimates the baseline using a median algorithm. It is based on the R function runmed(). Runmed is the ‘most robust’ scatter plot smoothing possible.
The estimation needs the number of iterations for the methods.
 Lab Tip! Lab Tip!
The baseline substraction is especially important for low molecular weight range.
It is strongly recommended to apply some baseline removal method when studying this mass range.
 Tech Hint! Tech Hint!
The baseline removal step uses the estimateBaseline() and removeBaseline() functions
of the MALDiquant package.
- Normalization:
This step calibrates the intensities of the mass spectra to equalize possible small batch effects. The user can choose one of the following methods: Total Ion Current (TIC), Probabilistic Quotient Normalization (PQN), and Median.
- TIC is a naïve method that calibrates all the mass spectra in their entire range using the total ion current (TIC) as normalization value.
- PQN uses the Dieterle et al. (2006) algorithm defined as follows. First, it calibrates all spectra using the "TIC" calibration, then calculates a median reference spectrum and the quotients of all intensities of the spectra with those of the reference spectrum. After that, it calculates the median of these quotients for each spectrum. Finally, it divides all intensities of each spectrum by its median of quotients.
- Median: The mass spectra are rescaled such that the median intensities are set to one.
 Lab Tip! Lab Tip!
Please note that the use of a standard molecule to be added to the sample and resulting in the spectrum is not currently supported.
 Tech Hint! Tech Hint!
The normalization step uses the calibrateIntensity() function
of the MALDiquant package.
- Averaging:
This step performs an average of the spectra when multiple replicates of each sample are present. At the end of the execution, it provides a single averaged mass spectrum per sample.
The user can choose one of the following methods:
- Mean: for each m/z values, the intensity is computed as the average of the intensities of the replicates for the same m/z.
- Median: for each m/z values, the intensity is computed as the median of the intensities of the replicates.
- Sum: for each m/z values, the intensity is computed as the sum of the intensities of the replicates.
 Lab Tip! Lab Tip!
Please note that you cannot use the Sum method when the number of replicates per sample is variable.
 Tech Hint! Tech Hint!
The averaging step uses the averageMassSpectra() function
of the MALDiquant package.
- Alignment:
This step has two different goals: estimating the noise and aligning the spectra by correcting the phase. A preventive Signal-to-Noise Ratio (SNR) should be provided. The alignment is obtained by estimating a reference sample using a suitable warping function to overcome the difference between the mass positions in the reference and in the sample of interest to match the peaks within a given tolerance and re-calibrate the mass positions. The tolerance is the maximum relative deviation of a peak position to be considered identical (must be multiplied by 10-6 for ppm).
For estimating the noise level, the user can choose one among the following methods:
- MAD: it estimates the noise of mass spectrometry data by calculating the median absolute deviation.
- Super Smoother: it estimates the noise of mass spectrometry data by calculating the Friedman's SuperSmoother (Friedman, 1984)
For aligning the mass spectra, the user can choose one among the following base wrapping functions:
- LOWESS: it uses the Local Weight Scatterplot Smoothing to re-calibrate the mass positions during the alignment.
- Linear estimation: it uses the linear approximation (polynomial of degree 1) to re-calibrate the mass positions during the alignment.
- Quadratic estimation: it uses the quadratic approximation (polynomial of degree 2) to re-calibrate the mass positions during the alignment.
- Cubic estimation: it uses the cubic approximation (polynomial of degree 3) to re-calibrate the mass positions during the alignment.
 Tech Hint! Tech Hint!
The alignment uses the alignSpectra() function
of the MALDiquant package.
- Peak identification, extraction and selection subsection:
- Detection:
This step produces the peak feature matrix, i.e., it extracts the peaks from each sample and forms a matrix with the peak positions (the mass m/z) and the peak intensities where all samples are evaluated.
A peak is a local maximum of the mass spectrum with an intensity above a user-defined noise threshold.
To perform this step, we apply different sub-steps such as detection, binning, and filtering. The detection sub-step aims at identifying potential peaks, the binning sub-step looks for similar peaks (mass) across the mass spectra and equalizes their mass, the filter sub-step aims to remove infrequent occurring peaks that might be due to noise.
For the detection sub-step, the necessary parameters are inherited from the alignment step.
For the binning sub-step, the user can choose among the following methods
- Strict: the new peak position is the mean mass of a bin.
- Relaxed: the mean mass of the highest peaks inside the window.
For the selection sub-step, the user must provide the parameter Coverage that denotes the proportion of samples that detect the peak for its inclusion. The peaks that are present in a percentage of samples lower than the coverage are removed.
 Lab Tip! Lab Tip!
Note that the choice of the parameter Coverage is relevant for the rest of the analysis since it acts as a variance/bias trade-off.
A large value of this parameter leads to a smaller number of features selected as significant peaks.
In general, this choice reduces the variance among samples but increase the bias.
A small value of this parameter leads to an increasing number of peaks, reducing the bias and increasing the variance.
The optimal choice is difficult in an unsupervised context, while it could be guided from the data in a supervised context.
Therefore, the users could consider trying several analyses using different choices of the parameter Coverage.
For example, if the spectra come from the same experimental condition, then it is suggested to choose a relatively high value for the Coverage to capture the samples' commonalities.
Instead, if the spectra come from two or more experimental conditions, it should be decreased (about proportionally to the less abundant class) to detect differences across conditions.
 Tech Hint! Tech Hint!
The detection step uses the detectPeaks(), binPeaks() and filterPeaks() functions
of the MALDiquant package.
- Clustering and visualization subsection:
- Clustering:
In this step, we first create a similarity matrix using the cosine correlation method on the feature matrix (peaks matrix). The similarity matrix is a symmetric matrix of dimension number of spectra x number of spectra. It contains the similarity among all possible pairs of spectra. Then, we apply a classical hierarchical clustering algorithm on the similarity matrix.
The user can choose among the following linkage function to create the dendrogram.
- Average: At each step and for each pair of clusters, it computes all pairwise distances between the spectra in the first cluster and the spectra in the second cluster. It considers the average of these distances as the distance between the two clusters, merging the two clusters with the minimum distance.
- Complete: At each step and for each pair of clusters, it computes all pairwise distances between the spectra in the first cluster and the spectra in the second cluster. It considers the maximum value of these distances as the distance between the two clusters, merging the two clusters with the minimum distance.
- Ward: At each step, it merges the pair of clusters with minimum between-cluster distance. In this way, it minimizes the total within-cluster variance.
- Median: At each step and for each pair of clusters, it computes all pairwise distances between the spectra in the first cluster and the spectra in the second cluster. It considers the median of these distances as the distance between the two clusters, merging the two clusters with the minimum distance.
The user can choose a pre-specified number of clusters (K value) to cut the dendrogram provide an expected number of clusters or can estimate such values from the data. If the user does not provide a possible number of clusters (K value), s/he can choose a method between the
- GAP statistic: the optimum number of cluster is estimated as the maximum of the GAP statistics (Tibshirani et al. 2001).
- Silhouette: the optimum number of cluster is estimated as the maximum of the average silhouette statistics (Rousseeuw, 1987).
 Tech Hint! Tech Hint!
The clustering uses the hclust() function from the stats package.
The Gap statistic is performed with the clusGap() function from the cluster package.
The silhouette method is performed with the cutree() function from the stats package and the silhouette() function from the cluster package.
- Heatmap:
This step allows creating a heatmap relative to the feature matrix (peaks matrix). The heatmap can be ordered according to clustering along one of its dimensions or both:
- None: the heatmap depicts the intensities in the order provided in the target files and the mass are ordered in an increasing way.
- Samples: the heatmap clusters the intensities by row (samples).
- Peaks: the heatmap clusters the intensities by column (peaks).
- Both: the heatmap clusters the intensities both by row and by column.
 Tech Hint! Tech Hint!
The heatmap step uses the pheatmap() function from pheatmap package.
- Principal Component Analysis (PCA):
This step allows creating the PCA projection of the feature matrix in the first three principal components' space. Then, it produces three plots comparing the scores of the first three components pairwise.
In these plots, it is possible to identify hidden structures among the samples. Moreover, if the target file contains a grouping variable, then it is possible to highlight such groups in the plots.
 Tech Hint! Tech Hint!
The PCA step uses the pca() function from the mixOmics package.
This section provides a summary of the job, including job and data set names, and steps, methods and parameters of the analysis.
A valid attribute file can be downloaded from here for later reuse.
| GeenaR_test job summary |
|---|
| User email | gabrielafernanda.coronelvargas@edu.unige.it | | Dataset | example | | Spectra list file | testTargetFile.txt | | Attributes file | GeenaR_test_attributes.csv | | Quality control | yes | | Reporting | yes, with code | | Trimming | 800 - 3000 | | Variance stabilization | yes | Method: sqrt | | | Smoothing | yes | Method: SavitzkyGolay | Half window size: 10 | | Baseline removal | yes | Method: SNIP | Number of iterations: 25 | | Normalization | yes | Method: TIC | | | Averaging | yes | Method: mean | | | Alignment | yes | Noise estimation: MAD
Phase correction: lowess | Half window size: 20, SNR: 2.0, Tolerance: 0.002 | | Peak detection | yes | Binning: strict
Selection |
Coverage: 0.2 | | Clustering | yes | Link function: average
K estimation: gap |
K value: 3 | | Heatmap | yes | Clustering: Samples | | | PCA | yes |
THis section provides information on the running job. Main steps are listed with starting and ending time.
At the end of the execution, link(s) to report(s) and the feature matrix are also provided, according to the request specified by the user.
| GeenaR_test elaboration and results |
|---|
GeenaR job GeenaR_test launched on January 28, 2021 at 11:17:59 UTC
Reading Mass Spectra START 11:18:03 399......... -- END 11:19:31 275
Acquiring Mass Spectra Metadata BEGIN 11:19:31 275 -- END 11:19:31 289
Saving RDS files BEGIN 11:19:31 289 -- END 11:19:33 310
READ DATA OK 11:19:33 310
Reading RDS files BEGIN 11:19:33 427 -- END 11:19:33 709
Writing Control Log File BEGIN 11:19:33 709 -- END 11:19:34 240
Plotting QC Pre-Trimming Plot BEGIN 11:19:34 240 -- END 11:19:38 268
Plotting Raw Mass Spectra BEGIN 11:19:38 269. -- END 11:19:51 086
QUALITY CONTROL PRE-TRIMMING OK 11:19:51 086
Reading RDS files BEGIN 11:19:51 140 -- END 11:19:51 421
Trimming Mass Spectra BEGIN 11:19:51 421 -- END 11:19:51 750
Saving RDS files BEGIN 11:19:51 750 -- END 11:19:53 114
Plotting Trimmed Mass Spectra BEGIN 11:19:53 114. -- END 11:20:05 062
TRIMMING OK 11:20:05 062
Reading RDS files BEGIN 11:20:05 120 -- END 11:20:05 278
Plotting QC Post-Trimming Plot BEGIN 11:20:05 279 -- END 11:20:08 454
QUALITY CONTROL POST-TRIMMING OK 11:20:08 454
Reading RDS files BEGIN 11:20:08 586 -- END 11:20:08 762
Variance Stabilization BEGIN 11:20:08 762 -- END 11:20:08 828
Plotting Stabilized Mass Spectra BEGIN 11:20:08 828.. -- END 11:20:35 904
Saving RDS files BEGIN 11:20:35 904 -- END 11:20:37 170
Smoothing BEGIN 11:20:37 170 -- END 11:20:37 412
Plotting Smoothed Mass Spectra BEGIN 11:20:37 412.. -- END 11:20:59 178
Saving RDS files BEGIN 11:20:59 178 -- END 11:21:00 596
Baseline Correction BEGIN 11:21:00 596 -- END 11:21:00 691
Plotting Corrected Mass Spectra BEGIN 11:21:00 691.. -- END 11:21:23 318
Saving RDS files BEGIN 11:21:23 318 -- END 11:21:24 760
Normalization BEGIN 11:21:24 760 -- END 11:21:24 900
Plotting Normalized Mass Spectra BEGIN 11:21:24 900.. -- END 11:21:48 347
Saving RDS files BEGIN 11:21:48 347 -- END 11:21:49 689
CLEANING MASS SPECTRA OK 11:21:49 689
Reading RDS files BEGIN 11:21:49 799
Loading Mass Spectra Metadata BEGIN 11:21:49 993 -- END 11:21:49 994
Averaging Mass Spectra Replicates BEGIN 11:21:49 994 -- END 11:21:50 220
Plotting Averaged Mass Spectra BEGIN 11:21:50 220 -- END 11:21:55 464
Aligning Mass Spectra BEGIN 11:21:55 464 -- END 11:21:55 617
Plotting Aligned Mass Spectra BEGIN 11:21:55 617 -- END 11:22:00 761
Saving RDS files BEGIN 11:22:00 761 -- END 11:22:01 413
ALIGNING MASS SPECTRA OK 11:22:01 413
Reading RDS files BEGIN 11:22:01 507 -- END 11:22:01 552
Detecting-Binning-Filtering Peaks BEGIN 11:22:01 552 -- END 11:22:01 661
Creating Feature Matrix BEGIN 11:22:01 661 -- END 11:22:01 709
Saving RDS files BEGIN 11:22:01 709 -- END 11:22:01 724
Plotting Mass Spectra Peaks BEGIN 11:22:01 724 -- END 11:22:02 983
PEAK EXTRACTION OK 11:22:02 983
Reading RDS files BEGIN 11:22:03 246 -- END 11:22:03 264
Plotting Principal Component Analysis BEGIN 11:22:03 264 -- END 11:22:05 716
Creating Distance Matrix BEGIN 11:22:05 716 -- END 11:22:05 731
Plotting Heatmap With Clustering BEGIN 11:22:05 731 -- END 11:22:06 452
Performing Gap Statistic BEGIN 11:22:06 452
Saving RDS files BEGIN 11:22:07 935 -- END 11:22:07 935
Plotting Dendrogram BEGIN 11:22:07 935
Saving RDS files BEGIN 11:22:08 747 -- END 11:22:08 747
CLUSTERING OK 11:22:08 747
REPORTING WITH MASS SPECTRA OK 11:22:22 504
REPORTING WITH CODE OK 11:22:24 340
Process completed on January 28, 2021 at 11:22:24 000
The result of the elaboration is now available at the following page: geenar_report_html.html.
The report including the R code is available at the following page: geenar_report_html_code.html.
The Feature Matrix is available at the following page: feature_matrix.csv. |
In the results section of the result page some of the main figures created during the run are reported in two tables.
The exact number and type of figures depends on the analysis steps selected by user for the run. Here we show all figures as they appears at the end of a 'complete' analysis.
The figures are shown with a reduced size. By clicking on any figure, it is opened in full size in a new window.
Figures can be downloaded by using the standard procedure: click on the figure with the right button of the mouse and then select the 'Save as' option..
The 'Quality control and Clustering' table reports the plot of the atypicality scores produced during the quality controls carried out both before and after the trimming
as well as the heatmap and the dendrogram generated by the Clustering steps..
| Quality control and Clustering |
Pre-trim quality control
 |
Post-trim quality control
 |
Peaks heatmap
 |
Dendrogram
 |
The 'Principal Components Analysis' table reports the partial loadings of the three most relevant components (PCA1, PCA2, PCA3) and the plot of all samples in two dimensional graphs.
The partial loading of each component includes the list of the 25 most relevant signals annotated with their relative weight for the component.
Plots includes group-related ellipses, supporting the visual interpretation of the effective separation of samples belonging to distinct groups.
| Principal Components Analysis |
PC1 loadings
 |
PC2 loadings
 |
PC3 loadings
 |
PC1 vs PC2
 |
PC1 vs PC3
 |
PC2 vs PC3
 |
An email message will be sent at the end of the run to the email address provided by the user.
The message will not include the results of the elaboration, but links to retrieve them.
Here below an example of the message is reported.
Subject: GeenaR GeenaR_test analysis from gabrielafernanda.coronelvargas@edu.unige.it (Italy)
A new job has been submitted to GeenaR!
User email: gabrielafernanda.coronelvargas@edu.unige.it
User country: Italy
GeenaR job GeenaR_test
Attributes file: http://proteomics.hsanmartino.it/geenar/run/GeenaR_test/attributes.csv
Target file: http://proteomics.hsanmartino.it/geenar/run/GeenaR_test/targetfile.txt
Report: http://proteomics.hsanmartino.it/geenar/run/GeenaR_test/Results/geenar_report_html.html
Report including code: http://proteomics.hsanmartino.it/geenar/run/GeenaR_test/Results/geenar_report_html_code.html
Feature Matrix: http://proteomics.hsanmartino.it/geenar/run/GeenaR_test/Results/feature_matrix.csv
References
- A.J. Hedges. A method to apply the robust estimator of dispersion, Qn, to fully-nested designs in the analysis of variance of microbiological count data. J Microbiol Methods. 2008, 72(2):206-207.
DOI: 10.1016/j.mimet.2007.11.021
- A. Savitzky and M. J. Golay. 1964. Smoothing and differentiation of data by simplified least squares procedures. Analytical chemistry, 36(8), 1627-1639.
DOI: 10.1021/ac60214a047
- M. U. Bromba and H. Ziegler. 1981. Application hints for Savitzky-Golay digital smoothing filters. Analytical Chemistry, 53(11), 1583-1586.
DOI: 10.1021/ac00234a011
- C.G. Ryan, E. Clayton, W.L. Griffin, S.H. Sie, and D.R. Cousens. 1988. Snip, a statistics-sensitive background treatment for the quantitative analysis of pixe spectra in geoscience applications. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms, 34(3): 396-402.
DOI: 10.1016/0168-583X(88)90063-8
- M. Morhac. 2009. An algorithm for determination of peak regions and baseline elimination in spectroscopic data. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment, 600(2), 478-487.
DOI: 10.1016/j.nima.2008.11.132
- M. van Herk. 1992. A Fast Algorithm for Local Minimum and Maximum Filters on Rectangular and Octagonal Kernels. Pattern Recognition Letters 13.7: 517-521.
DOI: 10.1016/0167-8655(92)90069-C
- J. Y. Gil and M. Werman. 1996. Computing 2-Dimensional Min, Median and Max Filters. IEEE Transactions: 504-507.
DOI: 10.1109/34.211471
- A. M. Andrew. 1979. Another efficient algorithm for convex hulls in two dimensions. Information Processing Letters, 9(5), 216-219.
DOI: 10.1016/0020-0190(79)90072-3
- F. Dieterle, A. Ross, G. Schlotterbeck, and Hans Senn. 2006. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Analytical Chemistry 78(13): 4281-4290.
DOI: 10.1021/ac051632c
- Friedman, J. H. (1984) A variable span scatterplot smoother. Laboratory for Computational Statistics, Stanford University Technical Report No. 5.
Web: scanned Department Copy
- Tibshirani, R., Walther, G. and Hastie, T. (2001). Estimating the number of data clusters via the Gap statistic. Journal of the Royal Statistical Society B, 63, 411–423.
Web: PDF
- Rousseeuw, P.J. (1987) Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math., 20, 53–65.
DOI: 10.1016/0377-0427(87)90125-7
|